195,00 € – 995,00 €
Product details
Synonyms = 58kDa Cytokeratin; CK5; Cytokeratin-5; DDD1; Epidermolysis Bullosa Simplex 2 (EBS2); Keratin 5; Keratin, Type II Cytoskeletal 5; Keratin-5; KRT5; Type-II Cytoskeletal 5; Type-II keratin Kb5
Antibody type = Recombinant Mouse monoclonal / IgG1
Clone = MSVA-605M
Positive control = Tonsil: A moderate to strong cytoplasmic KRT5 staining should be seen in virtually all squamous epithelial cells. Prostate: A strong cytoplasmic staining should be present in the majority of basal cells.
Negative control = Tonsil: Lymphatic cells must not show any KRT5 positivity.
Cellular localization = Cytoplasmic
Reactivity = Human
Application = Immunohistochemistry
Dilution = 1:100 – 1:200
Intended Use = Research Use Only
Relevance of Antibody
Cytokeratin 5 is expressed in basal cells and cancers derived from cytokeratin 5 expressing normal tissues.
Biology Behind
Cytokeratin 5 (CK5), also termed keratin 5 (KRT5) is a type II keratin protein encoded by the KRT5 gene on 12q13.13. It dimerizes with the type I keratin 14 and forms the intermediate filaments that primarily shape the cytoskeleton of basal epithelial cells. As other cytokeratins, KRT5 is part of the cytoskeletal scaffold within epithelial cells, which contributes to the cell architecture and provides the cells with the ability to withstand mechanical stress. Together with its partner protein keratin 14, KRT5 makes up for up to 25% of total cell protein in epidermal basal cells. Mutations of the KRT5 gene are leading to epidermolysis, an inherited skin blistering disease. The severity of the disease is linked to the position of the mutation within the protein. Expression of KRT5 (and 14) in cancer is indicating basal-like subtype which is linked to poor prognosis in several cancer types including breast and lung cancer.[1]
[1] Völkel et al. “Cytokeratin 5 and cytokeratin 6 expressions are unconnected in normal and cancerous tissues and have separate diagnostic implications” Virchows Archiv published online; September 24th 2021
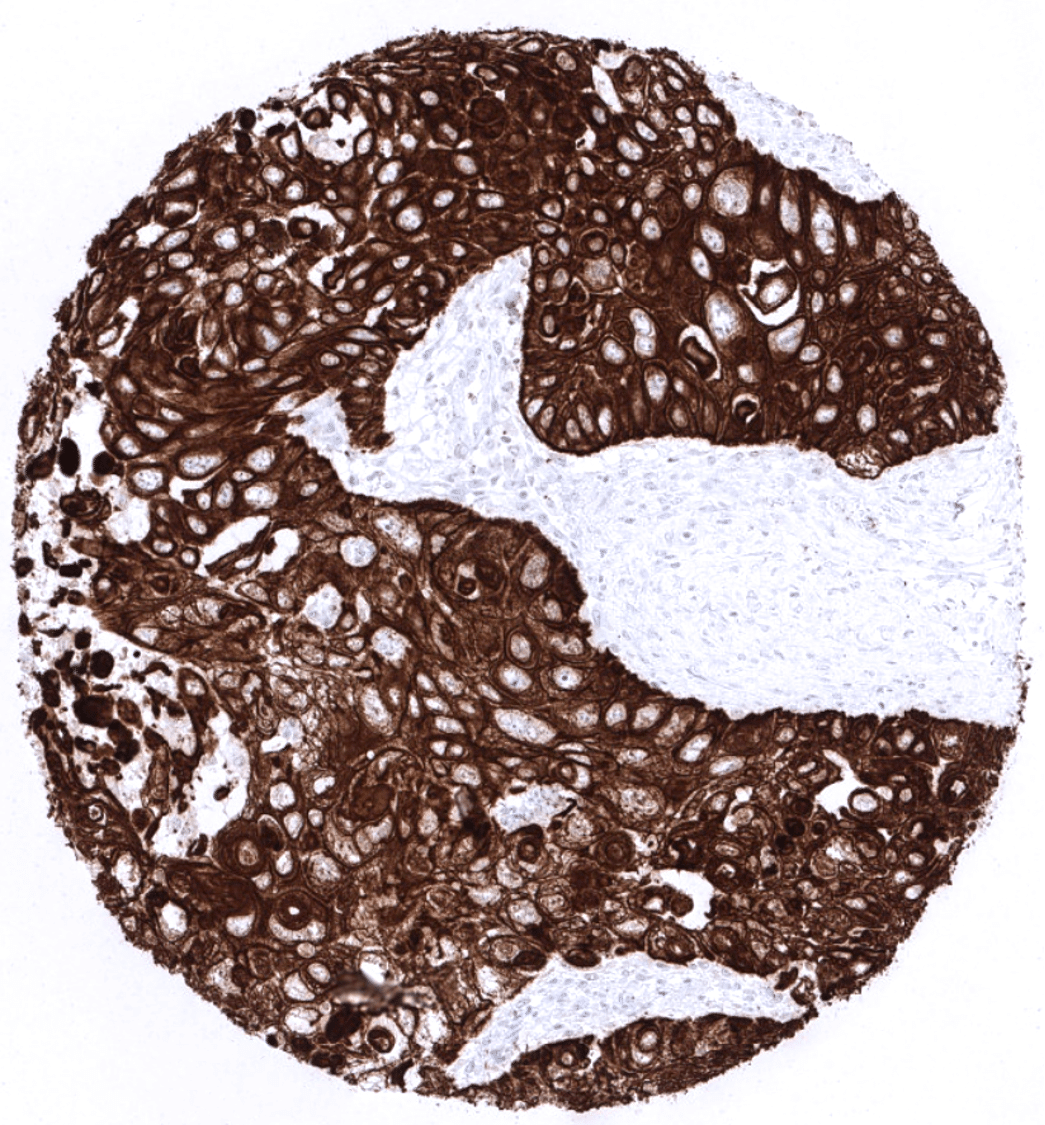
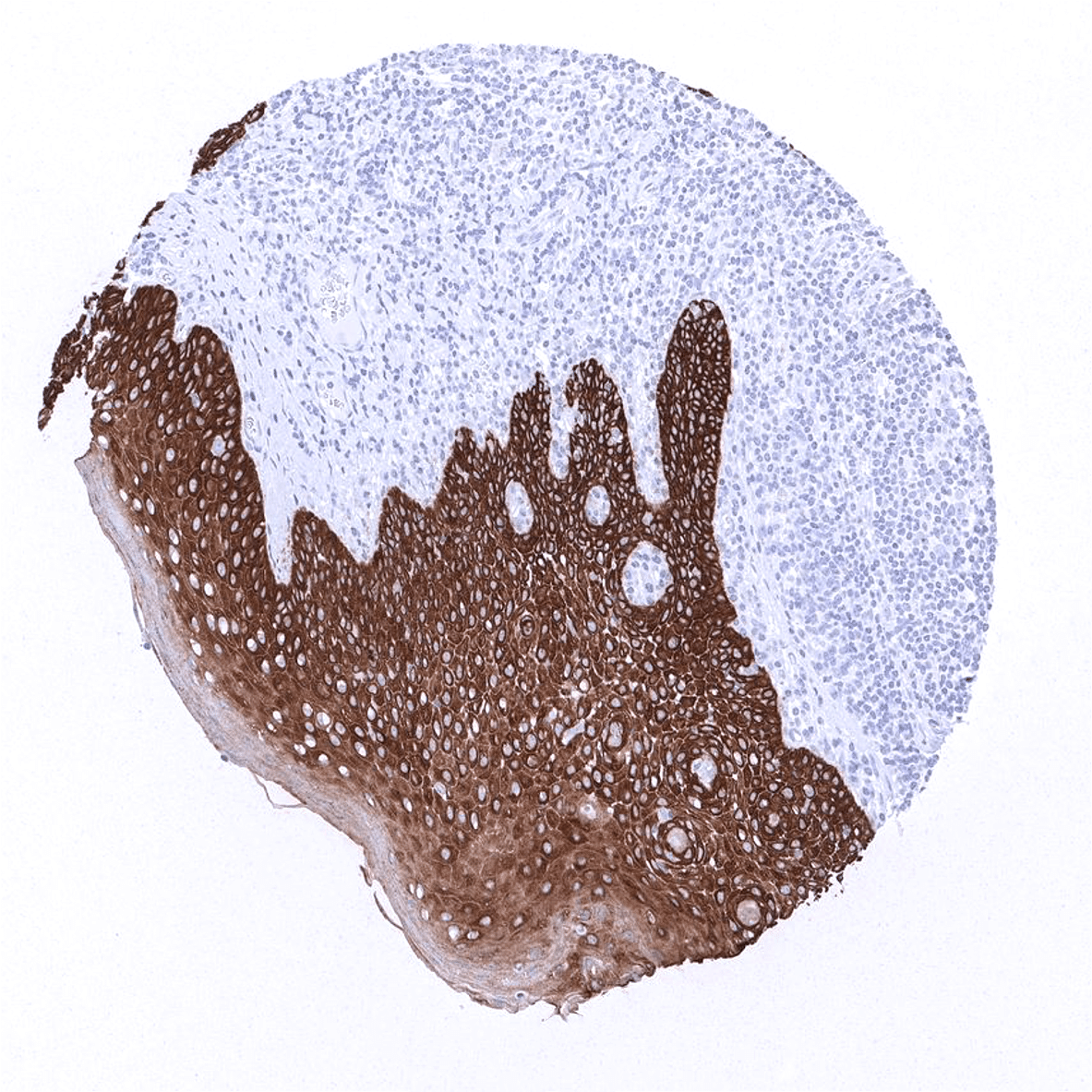
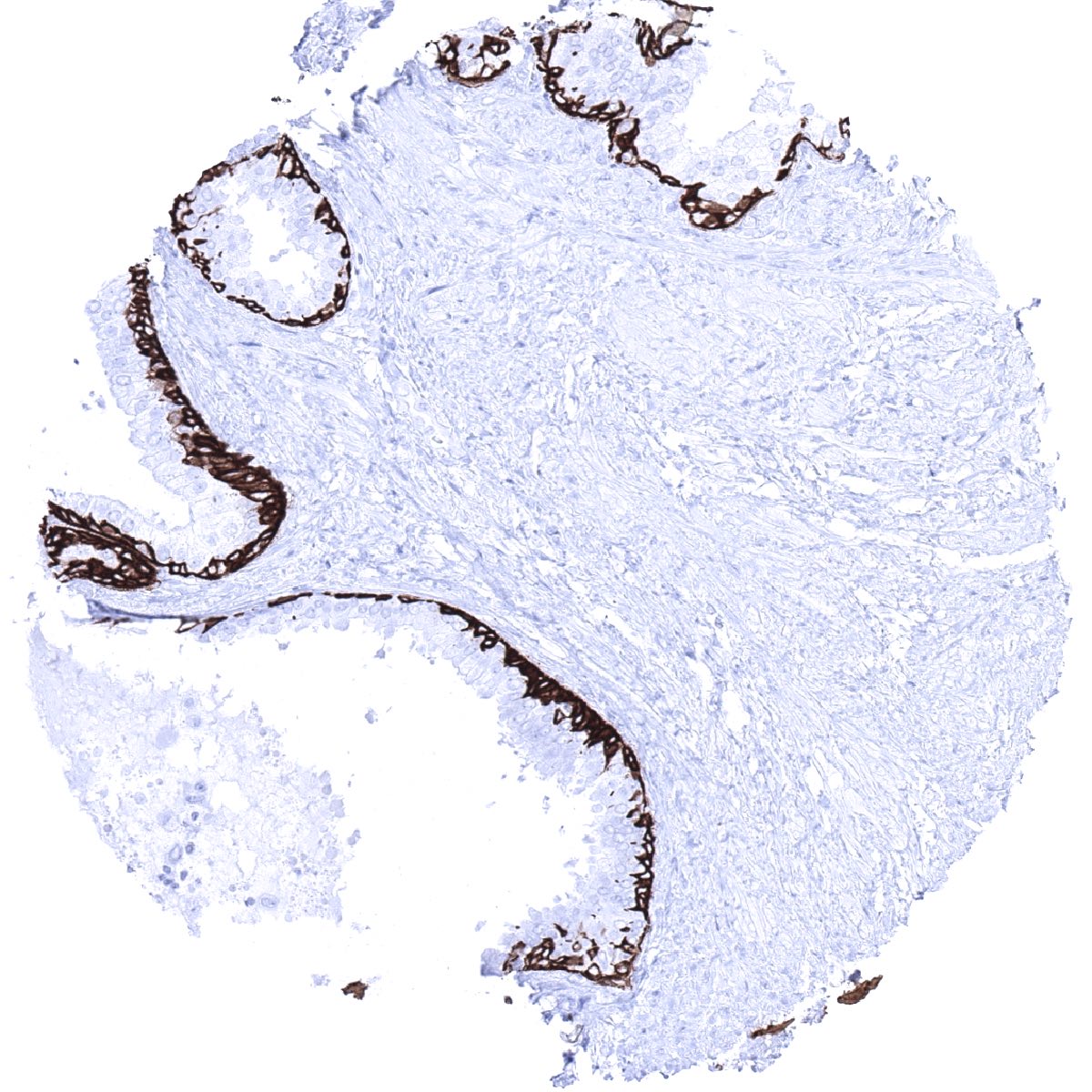
Staining Pattern in Normal Tissues
Cytokeratin 5 staining pattern in Normal Tissues with antibody MSVA-605M (images are shown in our “Normal Tissue Gallery”)
| Brain | Cerebrum | Negative. |
| Cerebellum | Negative. | |
| Endocrine Tissues | Thyroid | Negative |
| Parathyroid | Negative | |
| Adrenal gland | Negative. | |
| Pituitary gland | Negative. | |
| Respiratory system | Respiratory epithelium | Strong CK5 staining in basal cells of respiratory epithelium. Moderate CK5 staining inmyoepithelial cells and basal cells of excretory ducts of bronchial glands. |
| Lung | Negative. | |
| Gastrointestinal Tract | Salivary glands | Strong CK5 staining in myoepithelial cells and basal cells of excretory ducts. |
| Esophagus | Strong CK5 positivity of all squamous epithelial cell layers. Tendency to a slightly decreased staining towards the surface cell layers. | |
| Stomach | Negative. | |
| Duodenum | Negative. | |
| Small intestine | Negative. | |
| Appendix | Negative. | |
| Colon | Negative. | |
| Rectum | Negative. | |
| Liver | Negative. | |
| Gallbladder | Negative. | |
| Pancreas | Negative. | |
| Genitourinary | Kidney | Negative. |
| Urothelium | In the urothelium, only the basal cell layer stains CK5 positive. | |
| Male genital | Prostate | Strong CK5 staining in basal cells. |
| Seminal vesicles | Strong CK5 staining in basal cells. | |
| Testis | Negative. | |
| Epididymis | Strong CK5 staining of basal cells. A much weaker staining can be seen in a subset of columnar cells. | |
| Female genital | Breast | Strong CK5 staining in myoepithelial cells. |
| Uterus, myometrium | Negative. | |
| Uterus, ectocervix | Strong CK5 positivity of all squamous epithelial cells. Tendency to a slightly decreased staining towards the surface cell layers. | |
| Uterus endocervix | Strong CK5 staining in basal cells of some glands. | |
| Uterus, endometrium | Few endometrial epithelial cells may show CK5 positivity in some samples. | |
| Fallopian Tube | Negative. | |
| Ovary | Negative. | |
| Placenta early | Negative. | |
| Placenta mature | Negative. | |
| Amnion | Moderate to strong staining of amnion cells. | |
| Chorion | Moderate to strong staining of chorion cells. | |
| Skin | Epidermis | Strong CK15 staining in the lower half of the epidermis. Weaker or absent staining in the upper half of the squamous epithelium. |
| Sebaceous glands | Strong CK15 staining occurs in sebaceous glands and hair follicles. | |
| Muscle/connective tissue | Heart muscle | Negative. |
| Skeletal muscle | Negative. | |
| Smooth muscle | Negative. | |
| Vessel walls | Negative. | |
| Fat | Negative. | |
| Stroma | Negative. | |
| Endothelium | Negative. | |
| Bone marrow/ lymphoid tissue | Bone marrow | Negative. |
| Lymph node | Negative. | |
| Spleen | Negative. | |
| Thymus | Strong CK5 staining of epithelial cells including corpuscles of Hassall’s. | |
| Tonsil | Strong CK5 staining of squamous epithelial cells including tonsil crypts. | |
| Remarks |
These findings are largely comparable to the data described in the Human Protein Atlas (Tissue expression Cytokeratin 5). They also clarify that the RNA expression found in thymic tissue is caused by KRT5 expression of virtually all thymic epithelial cells.
Suggested positive tissue control: Tonsil: A moderate to strong cytoplasmic KRT5 staining should be seen in virtually all squamous epithelial cells. Prostate: A strong cytoplasmic staining should be present in the majority of basal cells.
Suggested negative tissue control: Tonsil: Lymphatic cells must not show any KRT5 positivity.
Staining Pattern in Relevant Tumor Types
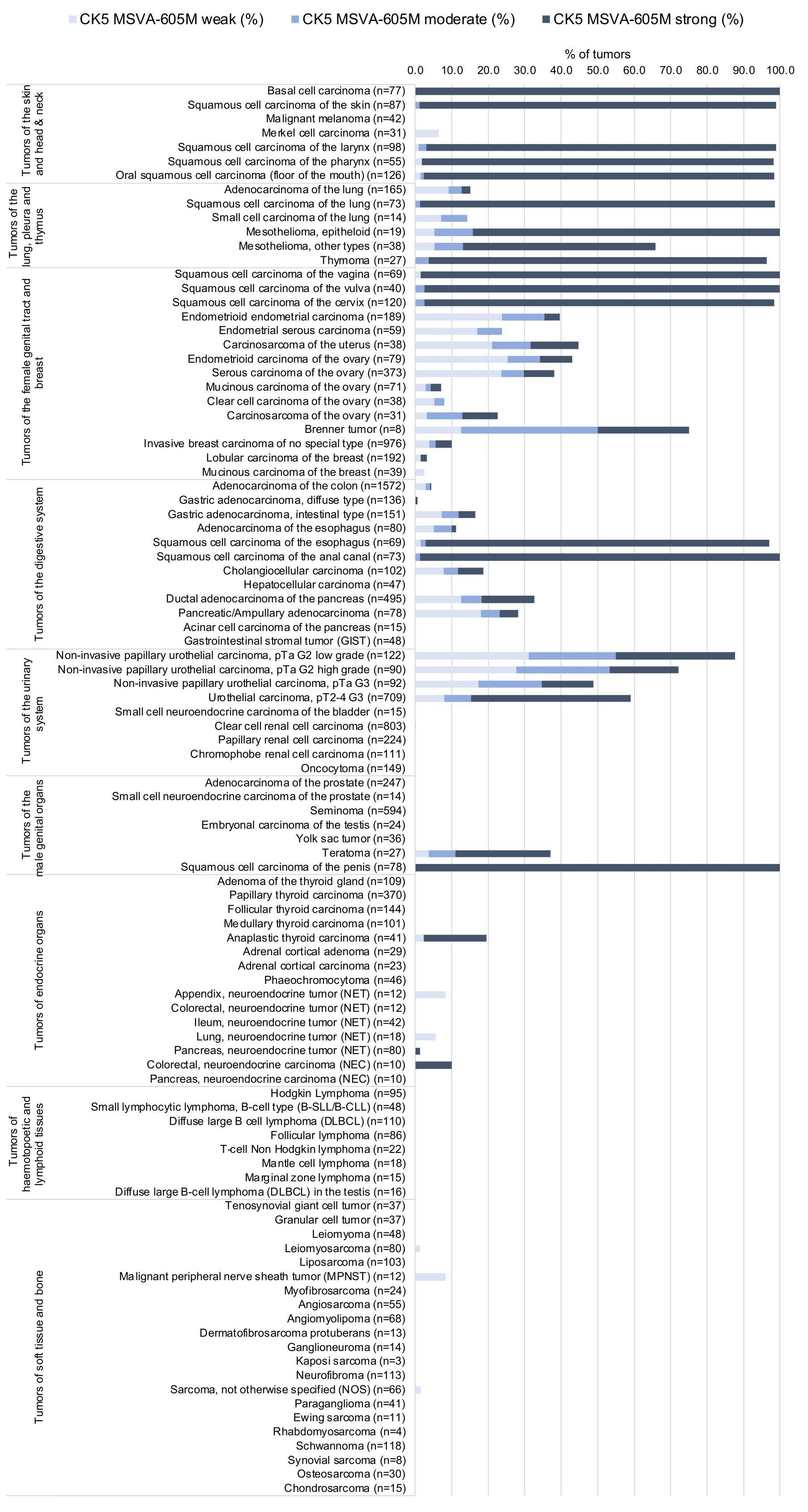
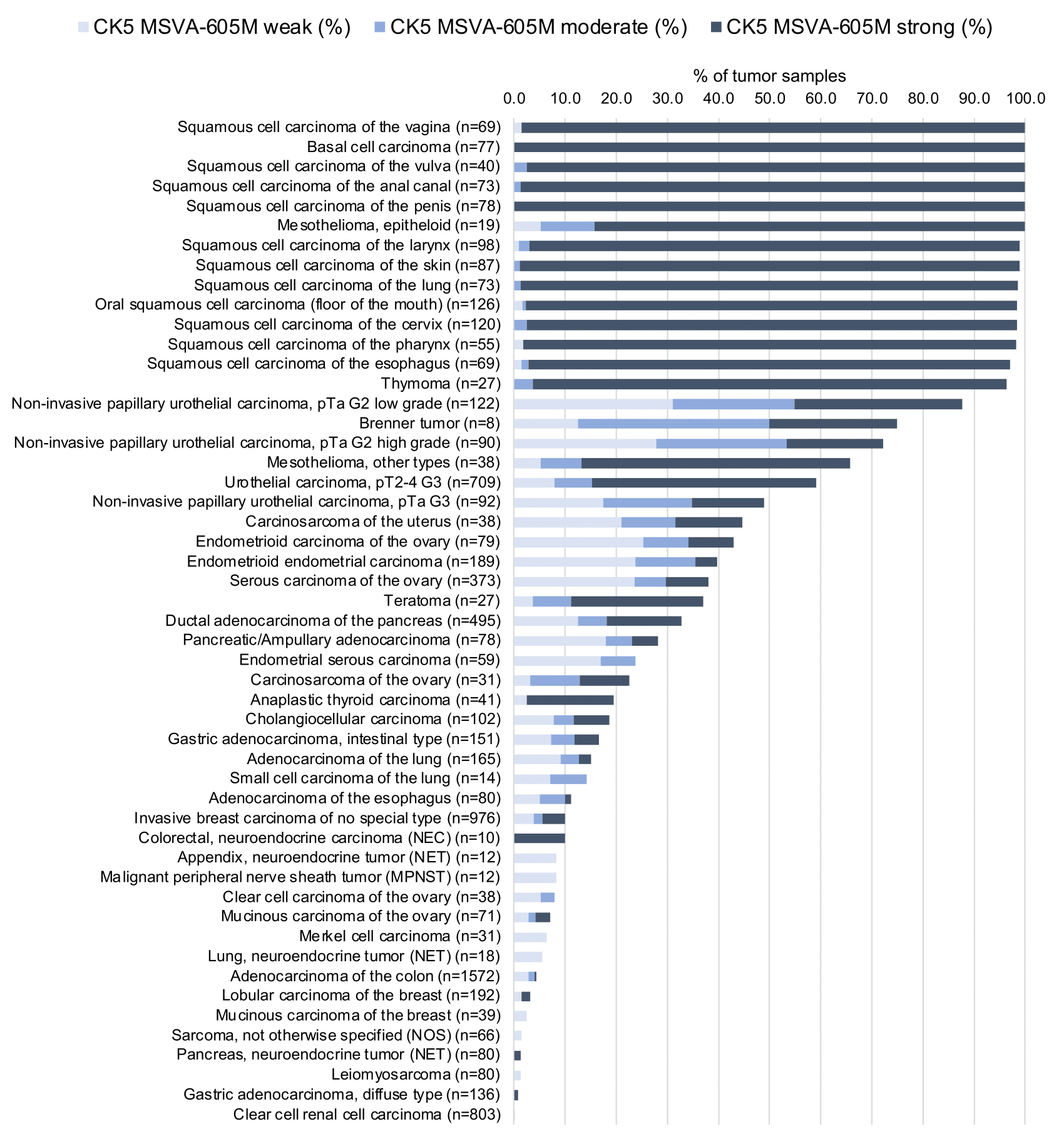
KRT5 expression is seen in almost all squamous cell carcinomas irrespective of their origin and differentiation. KRT5 is also seen in almost all thymic epithelial tumors, >80% of epitheloid type malignant mesothelioma, >60% of urothelial carcinomas of the urinary bladder, about 40% of endometroid carcinomas of the endometrium or ovary, 25-35% of pancreatic adenocarcinomas, 15-20% of cholangiocarcinomas of the liver, and in other cancers.
Detailed data on CK5 staining by MSVA-605M obtained from an analysis of 12,525 tumors from 120 different tumor types and subtypes have recently been published by Völkel et al. “Cytokeratin 5 and cytokeratin 6 expressions are unconnected in normal and cancerous tissues and have separate diagnostic implications”
The TCGA findings on Cytokeratin 5 RNA expression in different tumor categories have been summarized in the Human Protein Atlas.
Compatibility of Antibodies
Cytokeratin 5 (MSVA-605M) publication summary:
Relevant publication: Völkel et al. Cytokeratin 5 and cytokeratin 6 expressions are unconnected in normal and cancerous tissues and have separate diagnostic implications” Published in Virchows Arch. 2022 Feb;480(2) PMID: 34559291
In this study, 12’525 tumors were analyzed from 120 different tumor categories by using the following protocol: Heat-induced antigen retrieval for 5 minutes in an autoclave at 121°C in pH 9 Target Retrieval Solution buffer. MSVA-605M at a dilution of 1:150 at 37°C for 60 minutes. Visualization of bound antibody by the EnVision Kit (Dako, Agilent). This protocol was also used for all stainings depicted in our tumor and normal tissue galleries.
At least one case with a positive CK5 immunostaining was seen in 62 (51.7%) and at least one case with a strong CK5 immunostaining was seen in 48 (40%) of 120 tumor categories. The distribution of positive staining results is shown in an “organ-systematic” (Figure 1) and in a “ranking order” (Figure 2) below (images based on a compilation of data from (Völkel et al.). Results on associations with histopathological and clinical parameters of tumor aggressiveness are also summarized below (Table 1, based on data from Völkel et al.).
Authors conclusions on diagnostic utility of CK5 immunostaining with respect to the distinction of benign versus malignant (Völkel et al.)
- CK5 is useful for the visualization of basal cells in the prostate.
Authors conclusions on diagnostic utility of CK5 immunostaining with respect to the distinction of different tumor entities (Völkel et al.)
- Distinction of malignant epitheloid mesotheliomas (positive in 100%) from adenocarcinomas (positive in 12,5%) of the lung.
- Distinction of squamous cell carcinoma (positive in 98,7%) adenocarcinoma (positive in 12,5%) of the lung.
Authors conclusions on prognostic/predictive role of CK5 expression (Völkel et al.)
- CK5 expression is linked to poor grade, loss of ER/PR as well as “triple negativity” in breast cancer (p<0,0001 each).
- CK5 expression loss is linked to poor grade within pTa bladder tumors (p<0,0001).
Data from the publication: Völkel et al. “Cytokeratin 5 and cytokeratin 6 expressions are unconnected in normal and cancerous tissues and have separate diagnostic implications” Published in Virchows Arch. 2022 Feb;480(2) PMID: 34559291 Summarized in own graphics:
1. CK5 staining in tumors “organ-specific” with antibody MSVA-605M
2. CK5 staining in tumors “ranking order” by positivity with antibody MSVA-605M.
Protocol Recommendations
IHC users have different preferences on how the stains should look like. Some prefer high staining intensity of the target stain and even accept some background. Others favor absolute specificity and lighter target stains. Factors that invariably lead to more intense staining include higher concentration of the antibody and visualization tools, longer incubation time, higher temperature during incubation, higher temperature and longer duration of the heat induced epitope retrieval (slide pretreatment). The impact of the pH during slide pretreatment has variable effects and depends on the antibody and the target protein. Accordingly, multiple different protocols can generate identical staining results.
All images and data shown here and in our image galleries are obtained by the manual protocol described below. Other protocols resulting in equivalent staining are described as well.
Manual protocol
Freshly cut sections should be used (less than 10 days between cutting and staining). Heat-induced antigen retrieval for 5 minutes in an autoclave at 121°C in pH 9 Target Retrieval Solution buffer. Primary antibody specific against KRT5 clone MSVA-605M, dilution 1:150 is applied at 37°C for 60 minutes. Visualization of bound antibody by the EnVision Kit (Dako, Agilent) according to the manufacturer’s directions.
Agilent / Dako – Autostainer Link 48
Pretreatment in PT-Link for 30 minutes at 95°C (pH high); FLEX peroxidase blocking for 5 minutes (room temperature), MSVA-605M 1:150 for 20 minutes (room temperature), FLEX+ mouse/rabbit (LINKER) for 15 minutes (room temperature), horseradish peroxidase (HRP) for 20 minutes (room temperature), FLEX DAB+Sub-Chromo for 10 minutes (room temperature), FLEX hematoxylin for 5 minutes (room temperature).
These images reflect stainings by the protocol described above. It is of note that a comparable staining result can also be obtained by different protocols. In general, a longer pretreatment, a longer incubation time of the primary antibody, a higher antibody concentration, and a longer incubation time of FLEX+LINKER result in stronger staining, potentially at the cost of more background staining. Modifications of the protocol with a strengthening effect on staining intensity in combination with changes of other parameters that result in lower staining intensity can result in a comparable result as shown above.
Leica – BOND RX
Dewax at 72°C for 30 seconds; Pretreatment in Bond Epitope Retrieval Solution (ER2 – EDTA pH9) for 20 minutes at 100°C; Peroxidase blocking for 5 minutes (room temperature), MSVA-605M 1:150 for 15 minutes (room temperature), Post primary (rabbit anti mouse) for 8 minutes (room temperature), Polymer (goat anti rabbit) for 8 minutes (room temperature), mixed DAB refine for 10 minutes (room temperature), hematoxylin for 5 minutes (room temperature).
These images reflect stainings by the protocol described above. It is of note that a comparable staining result can also be obtained by different protocols. In general, a longer pretreatment, a longer incubation time of the primary antibody, a higher antibody concentration, a higher temperature during incubation, and a longer incubation time of Post primary and or the Polymer result in stronger staining, potentially at the cost of more background staining. Modifications of the protocol with a strengthening effect on staining intensity in combination with changes of other parameters that result in lower staining intensity can result in a comparable result as shown above.
Roche – Ventana Discovery ULTRA
Pretreatment for 64 minutes at 100°C (pH 8,4); CM peroxidase blocking for 12 minutes (room temperature), MSVA-605M 1:150 for 20 minutes at 36°C, secondary antibody (anti-mouse HQ) for 12 minutes at 36°C, anti-HQ HRP for 12 minutes at room temperature, DAB at room temperature, hematoxylin II at room temperature for 8 minutes, bluing reagent at room temperature for 4 minutes.
These images depict staining results obtained by the protocol described above. It is of note, that the Ventana machines generally require higher antibody concentrations than other commonly used autostainers because the antibodies are automatically diluted during the procedure. Various other protocols can result in an identical result as shown above. A longer pretreatment, a longer incubation time of the primary antibody, a higher antibody concentration, a higher temperature during incubation, and a longer incubation time of secondary antibody and or the anti-HQ HRP result in stronger staining, potentially at the cost of more background staining.
Impact of pH
For antibody MSVA-605M pH9 is optimal but pH7,8 results in only slightly weaker stainings. At pH6, KRT5 immunostaining is markedly reduced.
Potential Research Applications
- Visualization of basal cells and myoepithelial cells in multicolor IHC approaches.
- The potential diagnostic utility of KRT5 expression analysis (as compared to KRT5/6 analysis) needs to be investigated.
- The clinical significance of KRT5 expression needs to be evaluated in tumor types containing significant subgroups of KRT5 positive and negative tumors.
Evidence for Antibody Specificity in IHC
Specificity of MSVA-605M is documented by strong positive staining in cell types that are well documented to express KRT5 such as squamous epithelia of all origins and basal cells in the prostate in combination with the absence of staining in all tissues known to not express KRT5 including tissues notorious for non-specific IHC background such as kidney, colonic mucosa, and epidermis.
In a recent publication, the specificity of MSVA-605M was demonstrated in a comparison vs another independent antibody. Völkel et al. “Cytokeratin 5 and cytokeratin 6 expressions are unconnected in normal and cancerous tissues and have separate diagnostic implications”